I’ll never forget the moment I found out that my cousin had cancer. It’s one of those moments you remember exactly where you were and what you were doing. I was a freshman in college and on my way to take an exam for Introduction to Engineering class. I had received a voicemail from my grandmother. I listened and immediately by the low, tearful tone of my grandmother’s voice. “Your cousin has stage 4 alveolar cancer. It’s a very aggressive form of cancer and the doctors say that she might not live another year.” I had no idea what I wanted to do with my life, until that moment.
My current research project focuses on discovering more personalized treatments for embryonal rhabdomyosarcoma. Our goal is to define the major subtypes of embryonal rhabdomyosarcoma by using a dendrogram – a method used to group things together based on how similar they are. We’re grouping together samples that have a similar genetic makeup by looking at their DNA and RNA sequencing. This dendrogram is ever-changing. We’re constantly adding more samples from patients, mice, and dogs, as well as trying to identify genes that we could target in a set a of patients. A lot of what we’re realizing is that there’s still so much to learn.
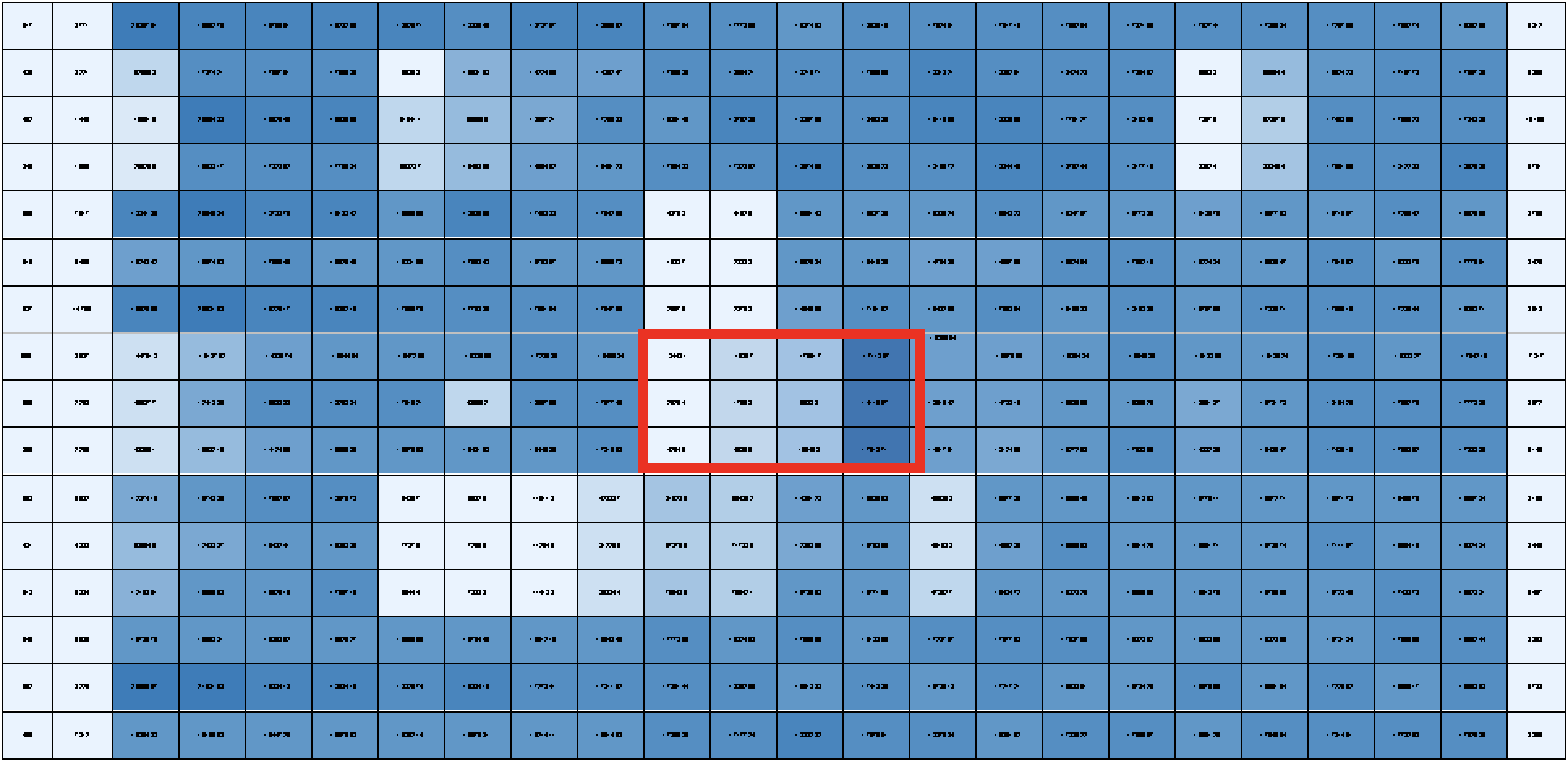
I’m also working on running drug screens on all the human and mouse cell lines included within the dendrogram. Our drug screens consist of a panel of 60 drugs with known activity in rhabdomyosarcoma. Each drug is tested at four different concentrations, and all in triplicate to ensure that the result we are seeing is real and not by chance. For example, one drug would be the what’s outlines in red, where the lighter blue represents greater cell death in a dose-dependent manner. Once the drug screen is completed for each group in the dendrogram we can look at synergistic effects of drugs – to see if two drugs would be more effective of stopping tumor growth than one.
Simultaneously, I’ve been learning about bioinformatics. Once we receive a sample from a patient, mouse, or dog we can get that sample sequenced. Perl and Linux are both useful tools I’m learning to better understand how to analyze these sequences. I’m also learning how to interpret the important mutations compared to others and what their function is within a tumor.
Finally, I’m very close to finishing two manuscripts that we’re hoping to publish- stay tuned! And check out the podcast Molly and I were on!
And for those who are wondering, my cousin is doing great!! She’s three years in remission and my family and I could not be more grateful. During my time at cc-TDI, I’ve been able to meet some pretty amazing people through our Nanocourse, when presenting my research, and visitors that come to tour our lab. I am tremendously grateful for the love and support that I’ve received and for the opportunity to make a difference in children’s lives.
I’d like to thank Consano and all of those who have graciously contributed to the funding of my project.


